🌍 Qatar University–Led Study, with UGA Collaboration, Demonstrates Rapid, Culture-Free Sepsis Detection Using SERS and AI
- Yiping Zhao
- Jan 3
- 3 min read
SuperRamanNet Achieves Near Clinical-Grade Accuracy in Real-World Validation
Researchers at Qatar University (QU), in close collaboration with the University of Georgia (UGA) and international clinical partners, have reported a major advance in rapid sepsis diagnostics. Published in Talanta, the study presents a culture-free, surface-enhanced Raman spectroscopy (SERS) and artificial intelligence–based platform capable of recognizing sepsis and identifying causative pathogens directly from blood with near clinical-grade accuracy.
Led by teams at Qatar University, this work reflects a strong QU–UGA research partnership that combines clinical access and large-scale validation in Qatar with UGA expertise in SERS substrates, plasmonic nanostructures, and AI-enabled spectral analysis.
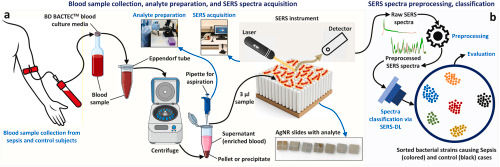
🩸 Rapid Blood-Based Diagnosis Without Culture
Conventional sepsis diagnostics rely heavily on blood culture, often requiring 24–72 hours to identify pathogens—time that critically ill patients may not have. In this QU-led study, blood samples from clinically confirmed sepsis patients and controls were collected at a tertiary hospital in Qatar and processed using a simple target-analyte enrichment protocol, avoiding microbial culture altogether.
Only microliter volumes of enriched blood were deposited onto silver nanorod (AgNR) SERS substrates, followed by rapid Raman spectral acquisition. This streamlined workflow enables direct vibrational fingerprinting of sepsis-causing pathogens from blood, making it well suited for point-of-care and near-patient testing.
🧠 SuperRamanNet: AI Designed for SERS and Clinical Translation
The SERS data were analyzed using SuperRamanNet, a lightweight one-dimensional deep learning model based on super operational neural networks (Super-ONNs). Developed within the QU–UGA collaboration, SuperRamanNet is specifically designed to handle the variability and complexity of biological SERS spectra.
Compared with conventional convolutional neural networks, SuperRamanNet incorporates nonlinear operations and learnable spectral shift-invariance, enabling more robust feature extraction. Despite this advanced architecture, the model remains compact and computationally efficient (~0.65 million parameters; ~0.0014 ms inference time per spectrum), supporting future deployment on portable Raman systems.
📊 Strong Clinical Performance and External Validation
Using SERS spectra from 653 analyte-enriched blood samples spanning five major sepsis-causing pathogens (Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Staphylococcus epidermidis) and negative controls, the QU-led team demonstrated:
99.67% accuracy for binary sepsis recognition
98.84% accuracy for six-class pathogen identification
Importantly, the system was tested on an independent external blind cohort, where it maintained 98.28% pathogen identification accuracy, underscoring its robustness and generalizability beyond the training data.
🔍 Interpretability and Confidence in AI Decisions
In addition to high accuracy, the study emphasizes interpretability, a critical requirement for clinical adoption. Feature-embedding visualizations (t-SNE and UMAP) revealed clear separation between pathogen classes, while saliency mapping identified key Raman bands driving classification.
These spectral regions align with known biochemical signatures of bacterial pathogens, helping build confidence in AI-assisted diagnostic decisions and supporting responsible clinical translation.
🤝 A Strong QU–UGA Partnership Toward Point-of-Care Sepsis Diagnostics
From the UGA perspective, this work highlights the value of international collaboration in translating advanced SERS and AI technologies toward real clinical impact. With Qatar University leading clinical design, data collection, and validation, and UGA contributing expertise in SERS substrates, plasmonics, and AI-driven spectral analysis, the partnership demonstrates how complementary strengths can accelerate innovation in biomedical diagnostics.
Future efforts will focus on expanding pathogen coverage, integrating clinical metadata, optimizing performance on portable Raman platforms, and advancing toward prospective clinical studies. Together, these steps bring SERS–AI–based sepsis diagnostics closer to real-world deployment, with the potential to significantly improve early detection and patient outcomes.
📄 Read the full paper:SERS on analyte-enriched blood for rapid, culture-free sepsis recognition and causative pathogen identification with super operational neural networks, Talanta (2026).





Comments